Research Interests
Evolution and ecology of the human gut microbiome
The human gut microbiome is a microcosm of complex microbial life, teeming with diverse bacterial communities. How these communities evolved for such a complex environment and respond to contemporary selection pressures is an open question. As just one example, the human gut harbors a plethora of antimicrobial resistance variation with potential to seed antibiotic resistance in opportunistic and obligate pathogens. However, it’s unknown how this ‘resistome’ is maintained, and with what rates and by what mechanisms horizontal gene transfer to pathogens is achieved (Van Schaik 2015). Using tools from evolutionary genetics, I study questions of adaptation like these in various human gut bacterial species. With this work, I hope to drive research to answer questions about microbiomes’ history, understand contemporary dynamics of selection, and build tools to shape the future of human health via gut communities. Preprint coming soon! :)
A scanning electron micrograph of Neisseria gonorrhoeae. Courtesy of the NIAID Rocky Mountain Laboratories in Hamilton, MT
Detecting adaptation under complex adaptive architectures
The capacity for adaptation beyond the classical hard sweep has been well-established, from soft sweeps at the same site to polygenic adaptation across sites. Methodology to detect adaptation has similarly evolved. However, our understanding of the dynamics and genetic architecture of adaptation under different evolutionary regimes is just scratching the surface. As signals of adaptation become both more subtle and difficult to interpret under polygenic adaptation, there is an exciting possibility to explore novel genomic signatures of adaptation, and enact new methodology made possible with greater computing power and genomic data (e.g. ARGs, haplotype-based statistics, frequency spectra, etc.). In a future project, I hope to build statistics with power to detect joint signals of adaptation across many sites in the genome sharing a selective pressure. Stay tuned!
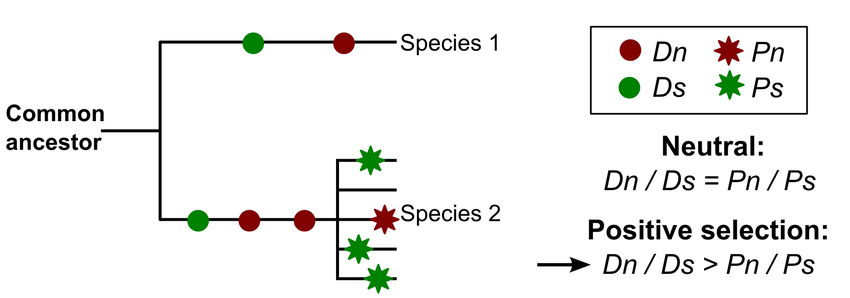
One of my favorite adaptation statistics from one of my favorite professors: the McDonald-Kreitman test
(Image from Salvador-Martínez 2016).
Statistical genetics of bacteria
Classic statistical genetics have (for the most part) been developed for populations of eukaryotic organisms, and there are few organisms so well-represented as bacterial species in classic experimental genetics. However, the plethora of bacterial data and a desire to map the genetic basis of heritable traits in bacteria have produced a lively field of bacterial statistical genetics. While interesting challenges remain in the mapping of non-single-site variation and ameliorating bacterial population structure for GWAS, there are a number of interesting applications for bacterial GWAS beyond the standard human repoitoire. In my undergraduate work at UChicago, I mapped the genetic basis of coinfection success for a dominant bacterial pathogen of Arabidopsis thaliana (preprint coming soon!). However, I believe the depth of bacterial sequences should allow the study of bacterial genetic architecture for different traits in natural populations of pathogens and other bacterial communities in great detail. Perhaps in the near future for me!
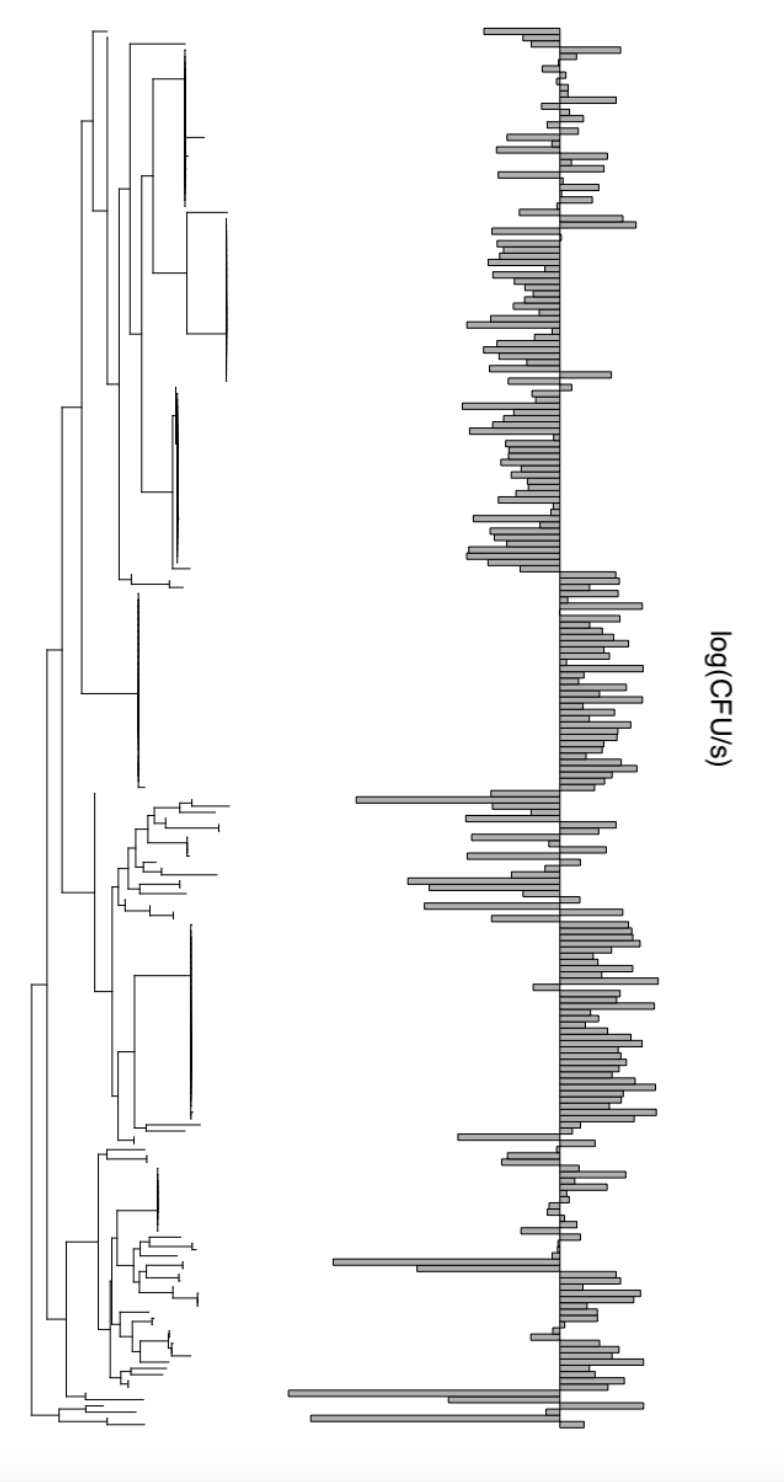
Luciferase counts for p23.A7, a dominant strain of the Arabidopsis pathogen Pseudomonas viridiflava. A figure for a future preprint? Oooh, how exciting!
References
Salvador-Martínez, Irepan. “Estimating complexity and adaptation in the embryo: a statistical developmental biology approach.” (2016).
Van Schaik, Willem. “The human gut resistome.” Philosophical Transactions of the Royal Society B: Biological Sciences 370.1670 (2015): 20140087.